Inductive Representation Learning on Large Graphs
(Hamilton et al., 2017)
Abstract
GraphSAGE is a framework for inductive representation learning on large graphs. Unlike transductive methods (which require all nodes during training), GraphSAGE generates node embeddings for unseen nodes by learning a function that samples and aggregates features from a node's local neighborhood.
The Problem with Early GNNs (Like GCN)
Early GNNs were Transductive.
- What that means: You have to feed entire graph into the training process.
- The limitation: You have to retrain the whole model to generate an embedding if new node is added, or some property changed.
- Scalability: You cannot easily process a graph with billions of nodes like (Pinterest or Facebook) because the whole adjacency matrix won't fit in GPU memory.
The GraphSAGE Solution
GraphSAGE (Graph Sample and Aggregate) introduced Inductive learning to graphs.
- What that means: Instead of learning embeddings for each node, GraphSAGE learns aggregating Functions.
- The Analogy:
- Transductive (GCN): Memorizing the map of New York City. If you go to London, you are lost.
- Inductive (GraphSAGE): Learning the skill of reading a map. You can now navigate New York, London, or a city that hasn't been built yet.
- High-Level Intuition: assumes that a node is defined by its neighbors
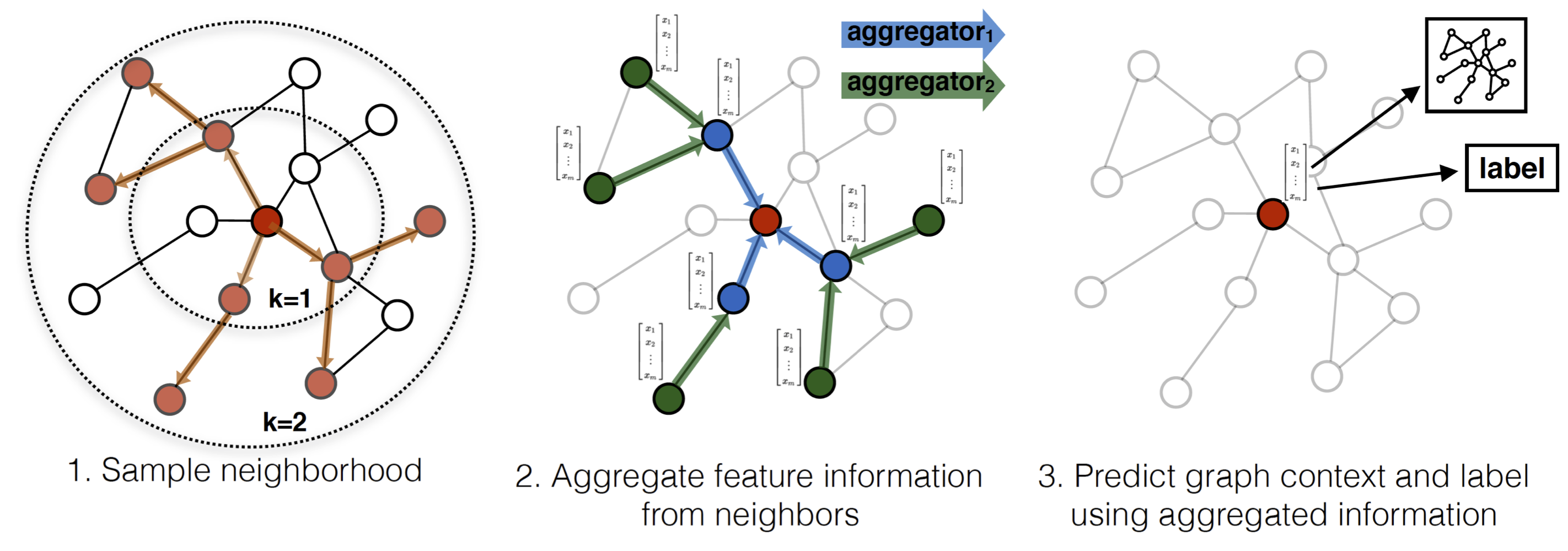
- Sampling: For a target node, select fixed number of neighbors (not all of them, just a sample). This keeps computation fixed regardless of node degree.
- Aggregating: Gather the feature information (text, images, stats) from those sampled neighbors and squash them together into a single vector.
- Updating: Combine the neighbors' aggregated info with the target node's current info to create a new embedding.
Method
The GraphSAGE algorithm generates embeddings by aggregating information from a node's local neighborhood. The process consists of three main steps: (1) Neighborhood Sampling, (2) Aggregation, and (3) Prediction/Loss.
- Forward Propagation Algorithm:
Let the graph be defined as $\mathcal{G} = (\mathcal{V}, \mathcal{E})$ with input features ${\{\mathbf{x}_v, \forall v \in \mathcal{V}\}}$. Let $K$ be the search depth (number of layers).We initialize the vector representations at $k=0$ as the input features: \[ \mathbf{h}^0_v = \mathbf{x}_v \]
For each layer $k = 1 \dots K$, and for each node $v \in \mathcal{V}$:
- Aggregate Neighbors: \[ \mathbf{h}^k_{\mathcal{N}(v)} = \text{AGGREGATE}_k \left( \{ \mathbf{h}^{k-1}_u, \forall u \in \mathcal{N}(v) \} \right) \]
- Update Node Embedding:
Combine the aggregated neighbor info with the node's own previous representation: \[ \mathbf{h}^k_v = \sigma \left( \mathbf{W}^k \cdot \text{CONCAT} \left( \mathbf{h}^{k-1}_v, \mathbf{h}^k_{\mathcal{N}(v)} \right) \right) \] where $\mathbf{W}^k$ is a learnable weight matrix and $\sigma$ is a non-linearity (e.g., ReLU). - Normalize: \[ \mathbf{h}^k_v = \frac{\mathbf{h}^k_v}{\| \mathbf{h}^k_v \|_2} \]
The final representation for node $v$ is $\mathbf{z}_v = \mathbf{h}^K_v$.
- Aggregation Functions:
The aggregation function must be permutation invariant (the order of neighbors should not matter). The paper proposes three distinct architecture choices:- Mean Aggregator:
This is the simplest approach. It takes the element-wise mean of the vectors in $\{ \mathbf{h}^{k-1}_u, \forall u \in \mathcal{N}(v) \}$. \[ \text{AGGREGATE}_k^{\text{mean}} = \frac{1}{|\mathcal{N}(v)|} \sum_{u \in \mathcal{N}(v)} \mathbf{h}^{k-1}_u \] Note: The paper notes that a variant of this (GCN-like) can concatenate the node $v$ with its neighbors before averaging, but the inductive Algorithm 1 keeps them separate during aggregation. - LSTM Aggregator:
LSTMs have higher expressive capability but are not naturally permutation invariant. To fix this, GraphSAGE adapts LSTMs to sets by applying them to a random permutation of the neighbors. \[ \text{AGGREGATE}_k^{\text{LSTM}} = \text{LSTM} \left( [ \mathbf{h}^{k-1}_{u_{\pi(1)}}, \dots, \mathbf{h}^{k-1}_{u_{\pi(|\mathcal{N}(v)|)}} ] \right) \] where $\pi$ is a random permutation function. - Pooling Aggregator:
This explicitly models the permutation invariance. Each neighbor's vector is passed through a fully connected neural network, followed by an element-wise max-pooling operation. \[ \text{AGGREGATE}_k^{\text{pool}} = \max \left( \{ \sigma \left( \mathbf{W}_{\text{pool}} \mathbf{h}^{k-1}_u + \mathbf{b} \right), \forall u \in \mathcal{N}(v) \} \right) \] where max denotes the element-wise maximum operator. This captures distinct features from the neighborhood (e.g., "is there any neighbor that is a fraudster?").
- Mean Aggregator:
- Neighborhood Sampling (Minibatching):
To keep the computational footprint manageable, GraphSAGE does not use the full set $\mathcal{N}(v)$. Instead, it uniformly samples a fixed size set of neighbors.Let $S_k$ be the sample size at layer $k$. If $|\mathcal{N}(v)| < S_k$, we sample with replacement; otherwise, we sample without replacement. This ensures the memory footprint per batch is fixed and predictable, regardless of the node degree.
- Loss Function:
GraphSAGE can be trained in a supervised or unsupervised manner.- Unsupervised (Negative Sampling):
We want nearby nodes to have similar embeddings, and disparate nodes to have distinct embeddings. For a node $u$, the loss function is: \[ J_{\mathcal{G}}(\mathbf{z}_u) = - \log \left( \sigma(\mathbf{z}_u^\top \mathbf{z}_v) \right) - Q \cdot \mathbb{E}_{v_n \sim P_n(v)} \left[ \log \left( \sigma(-\mathbf{z}_u^\top \mathbf{z}_{v_n}) \right) \right] \] where $v$ is a neighbor (positive sample), $P_n$ is a negative sampling distribution, and $Q$ is the number of negative samples. - Supervised:
For tasks like node classification, the unsupervised loss is replaced by a standard Cross-Entropy loss calculated on the final embeddings $\mathbf{z}_v$ of labeled nodes.
- Unsupervised (Negative Sampling):
Python
import sys
import torch
import torch.nn.functional as F
from torch_geometric.nn import SAGEConv
from torch_geometric.datasets import Planetoid
print(sys.version)
print(torch.__version__)
# 3.12.2 | packaged by conda-forge | (main, Feb 16 2024, 20:54:21) [Clang 16.0.6 ]
# 2.10.0
# Load cora citation data
dataset = Planetoid(root="/tmp/Cora", name="Cora")
data = dataset[0]
print(type(data))
#
# Graph Global Statistics
print(f"Nodes: {data.num_nodes}")
print(f"Edges: {data.num_edges}")
print(f"Average node degree: {data.num_edges / data.num_nodes:.2f}")
print(f"Isolated nodes: {data.has_isolated_nodes()}")
print(f"Self-loops: {data.has_self_loops()}")
print(f"Is undirected: {data.is_undirected()}")
# Feature and Label Analysis
print(f"Number of features: {dataset.num_features}")
print(f"Number of classes: {dataset.num_classes}")
# Class Distribution in Training Set
train_labels = data.y[data.train_mask]
values, counts = torch.unique(train_labels, return_counts=True)
for v, c in zip(values, counts):
print(f"Class {v.item()}: {c.item()} training samples")
# Connectivity Inspection
edge_index = data.edge_index
print(f"Edge Index Shape: {edge_index.shape}")
# Display first 5 edges
print(edge_index[:, :5])
"""
Output Context:
Nodes: 2708
Edges: 10556
Average node degree: 3.90
Isolated nodes: False
Self-loops: False
Is undirected: True
Number of features: 1433
Number of classes: 7
Class 0: 20 training samples
Class 1: 20 training samples
Class 2: 20 training samples
Class 3: 20 training samples
Class 4: 20 training samples
Class 5: 20 training samples
Class 6: 20 training samples
Edge Index Shape: torch.Size([2, 10556])
tensor([[ 633, 1862, 2582, 2, 652],
[ 0, 0, 0, 1, 1]])
"""
# Define GraphSAGE Model
class GraphSAGE(torch.nn.Module):
def __init__(self, in_channels, hidden_channels, out_channels):
super().__init__()
# Layer 1: Aggregates information from 1-hop neighbors
self.conv1 = SAGEConv(in_channels, hidden_channels, aggr='mean')
# Layer 2: Aggregates information from 2-hop neighbors (neighbors of neighbors)
self.conv2 = SAGEConv(hidden_channels, out_channels, aggr='mean')
def forward(self, x, edge_index):
# Step 1: Convolve (Aggregate neighbor info)
x = self.conv1(x, edge_index)
# Step 2: Activation (Add non-linearity)
x = x.relu()
# Step 3: Regularization (Prevent overfitting)
x = F.dropout(x, p=0.5, training=self.training)
# Step 4: Second Convolution
x = self.conv2(x, edge_index)
return F.log_softmax(x, dim=1)
# Initialize
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model = GraphSAGE(dataset.num_features, 64, dataset.num_classes).to(device)
data = data.to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=0.01, weight_decay=5e-4)
# Training loop
def train():
# 1. Prepare the model
model.train()
# 2. Reset Gradients
optimizer.zero_grad()
# 3. Forward Pass (The Prediction)
out = model(data.x, data.edge_index)
# 4. Calculate Loss (The Error)
loss = F.nll_loss(out[data.train_mask], data.y[data.train_mask])
# 5. Backward Pass (The Learning)
loss.backward()
optimizer.step()
return loss.item()
# Run it
for epoch in range(22):
loss = train()
print(f'Epoch {epoch}: Loss {loss:.4f}')
"""
Epoch 0: Loss 1.9501
Epoch 1: Loss 1.6858
Epoch 2: Loss 1.2758
Epoch 3: Loss 0.8277
Epoch 4: Loss 0.4822
Epoch 5: Loss 0.2450
Epoch 6: Loss 0.1192
Epoch 7: Loss 0.0669
Epoch 8: Loss 0.0340
Epoch 9: Loss 0.0196
Epoch 10: Loss 0.0093
Epoch 11: Loss 0.0098
Epoch 12: Loss 0.0036
Epoch 13: Loss 0.0028
Epoch 14: Loss 0.0012
Epoch 15: Loss 0.0023
Epoch 16: Loss 0.0013
Epoch 17: Loss 0.0012
Epoch 18: Loss 0.0005
Epoch 19: Loss 0.0023
Epoch 20: Loss 0.0011
Epoch 21: Loss 0.0004
""" References
- Hamilton, W. L., Ying, Z., & Leskovec, J. (2017). Inductive Representation Learning on Large Graphs. Advances in Neural Information Processing Systems (NeurIPS), 30.
- Kipf, T. N., & Welling, M. (2017). Semi-Supervised Classification with Graph Convolutional Networks. International Conference on Learning Representations (ICLR).